Estimating Fluxes in C3 Plants by Transient Labeling with Carbon-13 Labeled Carbon Dioxide - Part I: Objectives, Procedures and Results
steemstem·@davidrhodes124·
0.000 HBDEstimating Fluxes in C3 Plants by Transient Labeling with Carbon-13 Labeled Carbon Dioxide - Part I: Objectives, Procedures and Results
If you have read any of my recent posts you will have noticed that I am interested in plant metabolism and how to quantify (using computer simulation models) the rates of flow of material through metabolic pathways (fluxes). This has @steemstem relevance because it combines science, technology and mathematics and has applications in metabolic engineering ([Metabolic Engineering and Metabolic Flux Analysis](https://steemit.com/steemstem/@davidrhodes124/metabolic-engineering-and-metabolic-flux-analysis)). I am particularly interested in amino acid metabolism in plants and how the carbon skeletons of amino acids are derived via the central carbon metabolism networks that I have discussed previously ... the Calvin cycle of C3 photosynthesis ([The Calvin Cycle of C3 Photosynthesis](https://steemit.com/steemstem/@davidrhodes124/the-calvin-cycle-of-c3-photosynthesis), [Predicted Labeling Of Calvin Cycle Intermediates With Carbon-13 Labeled Carbon Dioxide](https://steemit.com/steemstem/@davidrhodes124/predicted-labeling-of-calvin-cycle-intermediates-with-carbon-13-labeled-carbon-dioxide)), photorespiration ([Photorespiration - Biochemistry](https://steemit.com/steemstem/@davidrhodes124/photorespiration-biochemistry)), glycolysis ([Glycolysis](https://steemit.com/steemstem/@davidrhodes124/glycolysis)), and the citric acid cycle ([Citric Acid Cycle and Mitochondrial Electron Transport](https://steemit.com/steemstem/@davidrhodes124/citric-acid-cycle-and-mitochondrial-electron-transport), [Citric Acid Cycle Labeling In C3 Plants](https://steemit.com/steemstem/@davidrhodes124/citric-acid-cycle-labeling-in-c3-plants)). Because the vast majority of plants use CO<sub>2</sub> from the atmosphere as their sole carbon source (*there are a few exceptions, such as parasitic plants*), it is possible to use stable-isotope labeled <sup>13</sup>CO<sub>2</sub> to monitor these fluxes during a time-course (transient labeling). Key information needed in this method of interpreting labeling patterns of intermediates in plants supplied with <sup>13</sup>CO<sub>2</sub> are pool sizes of as many intermediates that can be measured, and their changes in isotopic abundances over time (Morgan and Rhodes (2002)). This relies extensively on accurate quantification of multiple metabolites, and sensitive methods for determining their masses. Gas chromatography-mass spectrometry (GC-MS) is an excellent piece of technology to accomplish this. I would here like to illustrate these concepts using the model C3 plant *Arabidopsis thaliana* supplied with <sup>13</sup>CO<sub>2</sub>. **The following results are unpublished data as they essentially represent a feasibility study with the following objectives:** **Objectives** - Obtain a preliminary time-course to assess patterns of labeling of amino acids and pool size changes in the transition to photorespiratory conditions (~ 350 ppm to ~ 280 ppm of <sup>13</sup>CO<sub>2</sub>). - Acquire a data set for assessing precursor-product relationships in amino acid metabolism (e.g. Glyine --> Serine, Aspartate --> Asparagine, Aspartate --> Threonine, Glutamate --> Glutamine, Glutamate --> GABA, Glutamate --> Proline). - Optimize amino acid extraction, purification and derivatization procedures. - Optimize chromatography, especially concentrations of amino acid derivatives submitted for GC-MS analysis to minimize detector saturation. - Develop a computer simulation model to interpret time-courses of labeling of amino acids and amino acid pool size changes associated with transition of plants to photorespiratory conditions. This first post (Part I) will focus on experimental methods (procedures) and results. In Part II (to follow) I will discuss the integration of all the results into a computer simulation model. **Methods/Procedures** Wildtype, soil grown plants of *Arabidopsis thaliana* pre-grown under atmospheric CO<sub>2</sub> levels of approx. 350 ppm CO<sub>2</sub>, were supplied with sub-ambient <sup>13</sup>CO<sub>2</sub> (averaging approx. 280 ppm) for 9 h in an illuminated growth chamber. Samples were removed from the chamber at 1.5 h intervals and approximately 0.2 gFw (grams Fresh weight) of leaf tissue was extracted in 10 mL methanol. To each sample was added 250 nmol alpha-aminobutyrate (Aab) and alpha-aminoadipic acid (Aad) as internal standards. Samples were phase-separated by addition of 5 mL chloroform + 6 mL water. The upper aqueous phase was removed, dried under a stream of air, redissolved in 1 mL water and applied to freshly prepared columns (4 cm x 1 cm) of Dowex-1-acetate. The water wash from Dowex-1-acetate (7 mL total, containing neutral and basic amino acids) was applied to freshly prepared columns (4 cm x 1 cm) of Dowex-50-H<sup>+</sup>. The water wash from this column (approximately 13 mL) was discarded and neutral (plus basic) amino acids were eluted with 6 mL 6 M NH<sub>4</sub>OH. The latter fractions were dried under a stream of air. Acidic amino acids were eluted from Dowex-1-acetate with 6 mL 2 M acetic acid, the eluates dried under a stream of air, redissolved in 1 mL water and applied to freshly prepared columns (4 cm x 1 cm) of Dowex-50-H<sup>+</sup>. The water wash from this column (approximately 7 mL) was discarded and acidic amino acids were eluted with 6 mL 6 M NH<sub>4</sub>OH. The latter fractions were dried under a stream of air. An amino acid standard mixture (Sigma Chemical Co.) spiked with equimolar amounts of the internal standards, and the dried neutral (plus basic) and acidic amino acid samples were redissolved on 0.6 mL 60 % methanol, transferred to 1 mL microreaction vessels and evaporated to dryness under a stream of air, and derivatized to heptafluorobutryryl isobutyl (HFBI) derivatives. After drying, 0.2 mL methylene chloride was added and the solvent evaporated to dryness under a stream of air. To each sample 0.2 mL of a freshly prepared, chilled, 5 : 1 v/v mixture of isobutanol : acetyl chloride was added. Reaction vials were capped with cap and teflon coated septa, vortexed and heated at 120<sup>o</sup>C for 20 min in a pre-heated heating block. After 20 min, vials were removed from the heating block and allowed to cool to room temperature, and evaporated to dryness under a stream of air. After drying, 0.1 mL of heptafluorobutyric anhydride was added to each sample (in the fume hood). Caps and teflon-coated septa were replaced, samples were mixed, heated at 120<sup>o</sup>C for 10 min, allowed to cool to room temperature, and evaporated to the point of dryness under a stream of air. The first 4 samples in the time-course (t = 0 h, 1.5 h, 3.0 h, 4.5 h) were redissolved in 0.05 mL of ethyl acetate : acetic anhydride (1 : 1 v/v) and 1 uL aliquots analyzed by GC-MS using a Finnigan GCQ tandem mass spectrometer equipped with a db1 capillary column. Because results from these samples revealed detector saturation for the most abundant peaks (especially Asn and Gln in the neutral fraction, and Asp, Glu and Aad in the acidic fraction) the remaining 3 samples in the time-course (t = 6.0 h, 7.5 h, 9.0 h) were redissolved in 0.2 mL of ethyl acetate : acetic anhydride (1 : 1 v/v) and 1 uL aliquots analyzed by GC-MS (i.e. samples were diluted 4x). All samples were analyzed in electron ionization (ei) mode using the mass range 210 to 370 atmic mass units (amu). The amino acid derivatization methods described above was originally developed by MacKenzie and Tenaschuk (1974), and subsequently adapted by Siezen and Mague (1977). Our laboratory then adopted this method for analysis of amino acids in *Lemna minor* (Brunk and Rhodes (1988), Rhodes et al. (1981), (1986a), (1987), (1989)) and tissue cultured cells (Mayer et al. (1990), Rhodes et al. (1986b)). Amino acid extraction and ion exchange purification methods described above for *Arabidopsis* were adapted from Rhodes et al. (1981) and (1986b). **Amino acid abbreviations used** Aab = alpha-aminobutyrate (internal standard) Aad = alpha-aminoadipic acid (internal standard) Ala = alanine Gly = glycine Ser = serine Val = valine Thr = threonine Leu = leucine Ileu = isoleucine GABA = gamma-aminobutyrate Pro = proline Pglu = pyroglutamate Met = methionine Glu = glutamate Gln = glutamine Phe = phenylalanine Orn = ornithine Tyr = tyrosine Lys = Lysine His = Histidine **Results** *Chromatograms*. Shown below are GC-MS chromatograms (total ion intensities) of an amino acid standard mixture (used for determination of response factors of amino acids relative to the internal standards ((Aab) and (Aad)), and the neutral + basic and acidic amino acid fractions of *Arabidopsis* over the 9 h time course of feeding with <sup>13</sup>CO<sub>2</sub> (**asterisks denote detector saturation**): 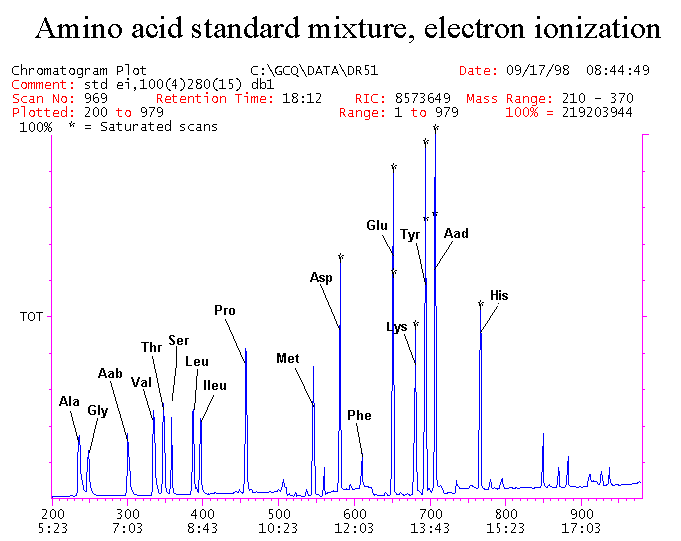 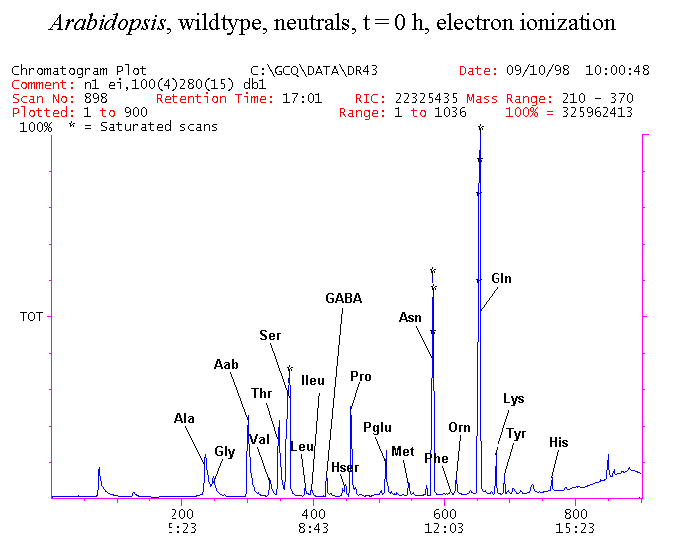 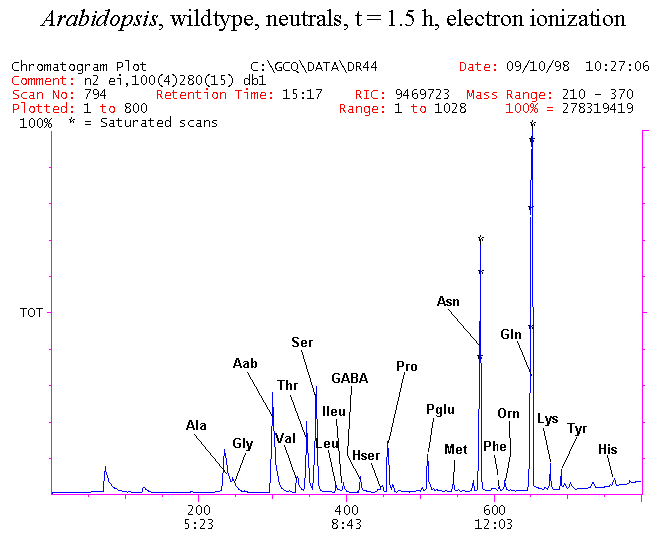 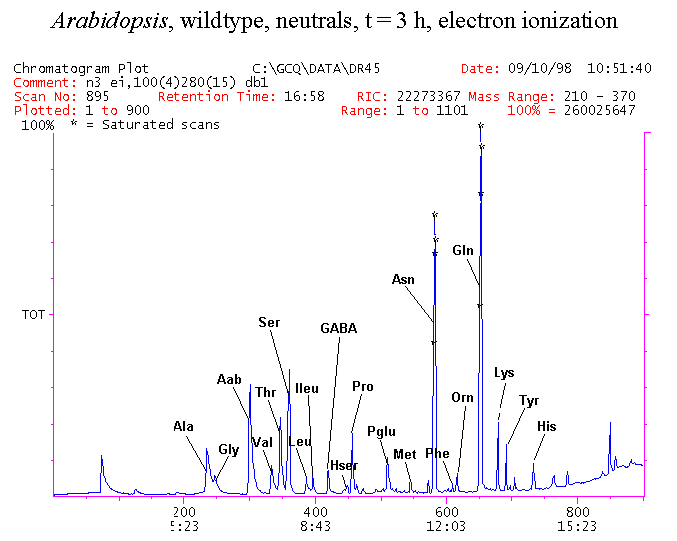 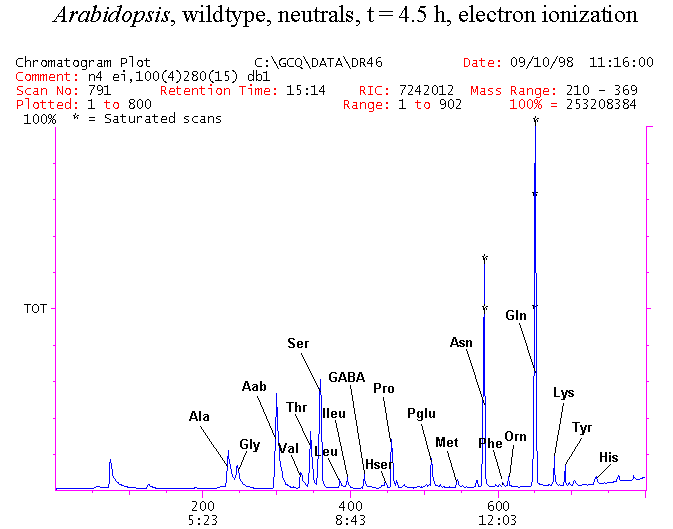 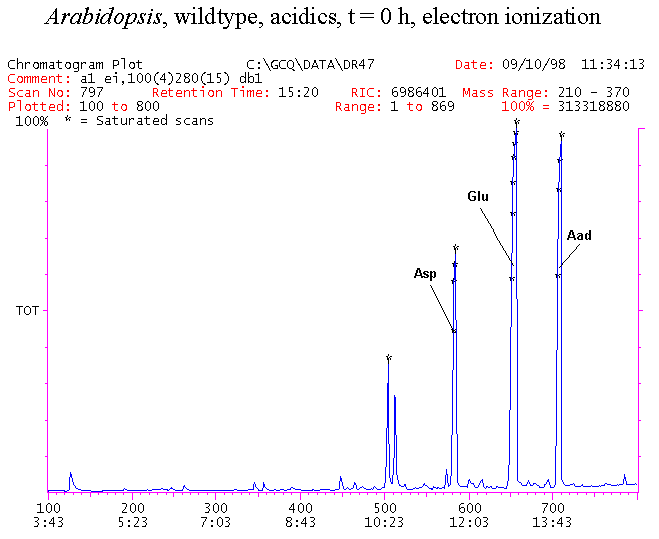 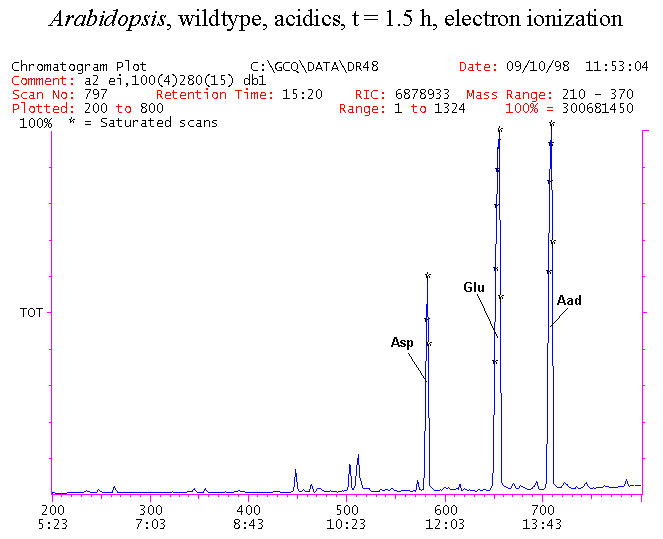 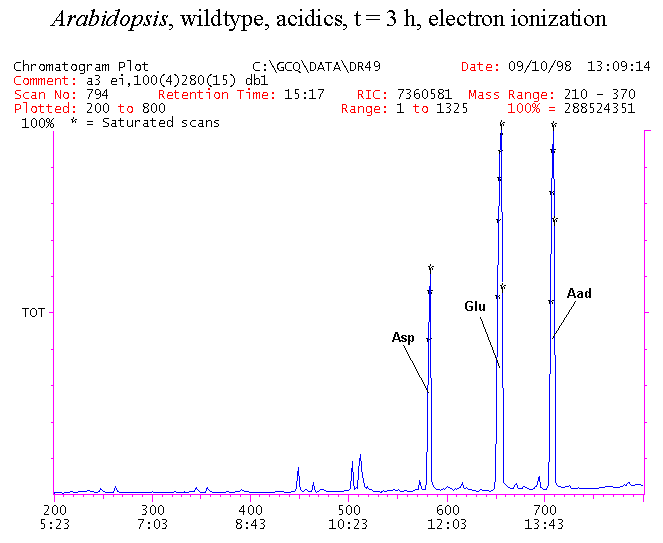 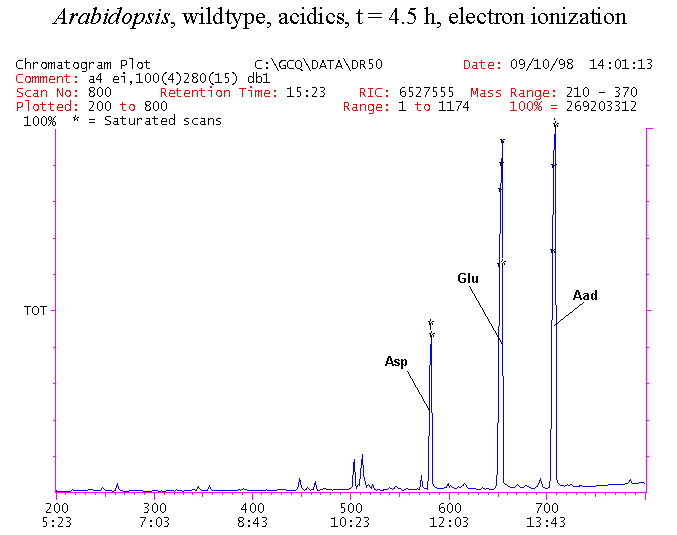 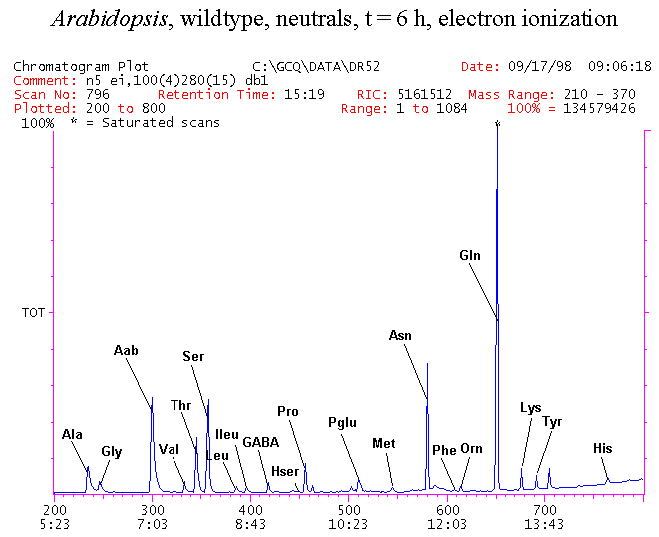 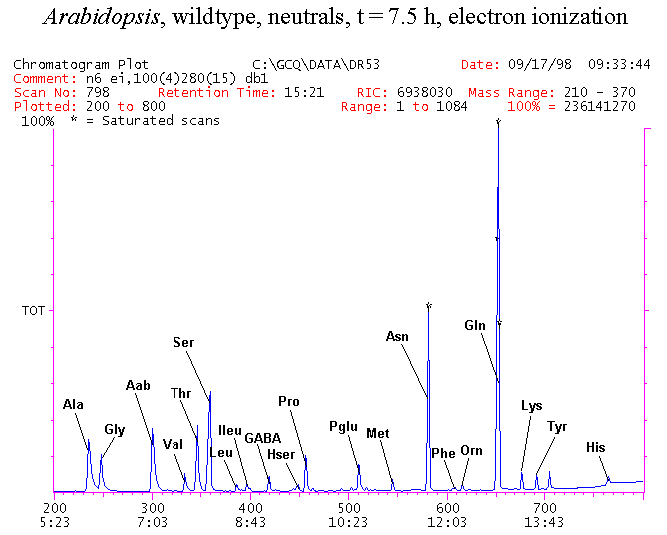 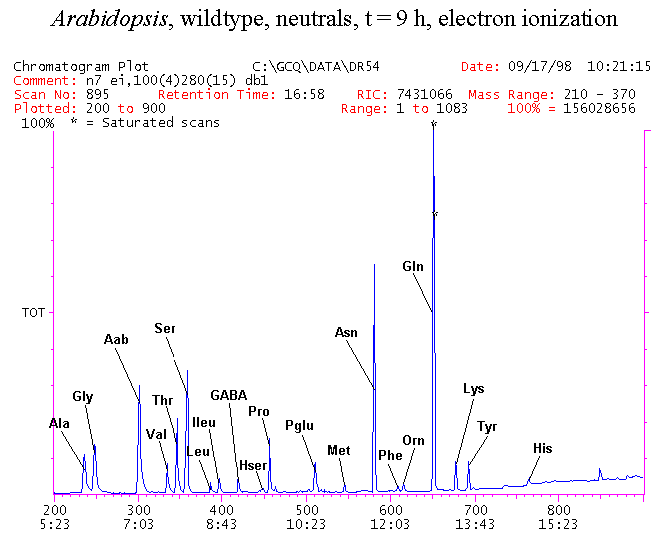 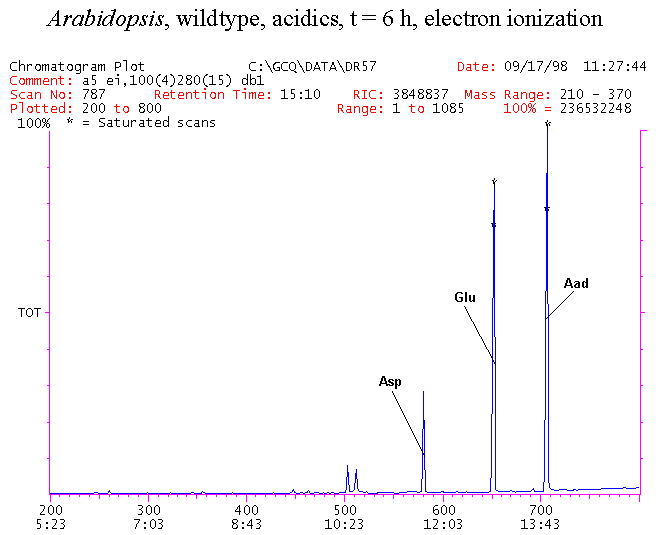 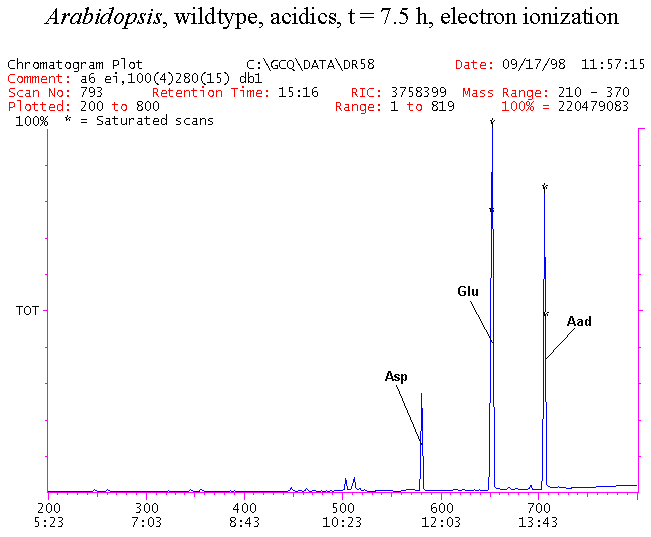 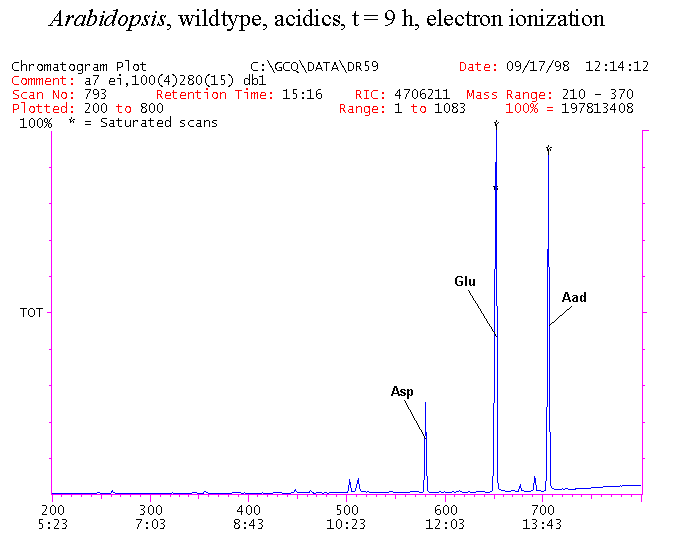 *Quantification of Pool Sizes*. The following Table summarizes the pool sizes of all of the amino acids detected in the above chromatograms, using the area counts under each peak and the amino acid standard mixture response factors for each amino acid relative to the internal standards (Aab and Aad): 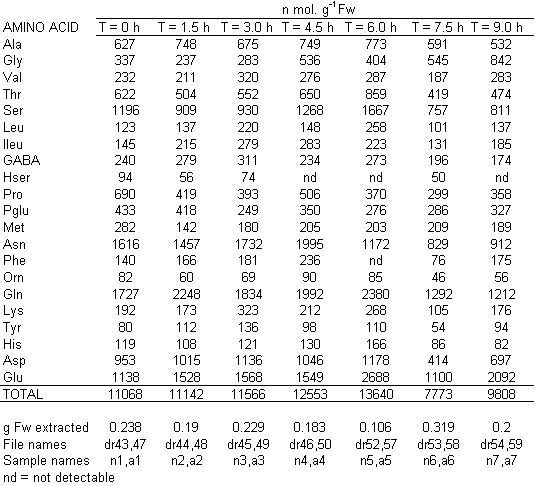 Structures of the N(O,S)-heptafluorobutyryl isobutyl (HFBI) derivatives of these amino acids are shown below. Each carboxyl group receives an isobutyl moiety (red), and each amino group, hydroxyl group (or free -SH group) receives a heptafluorobutyryl moiety (blue): 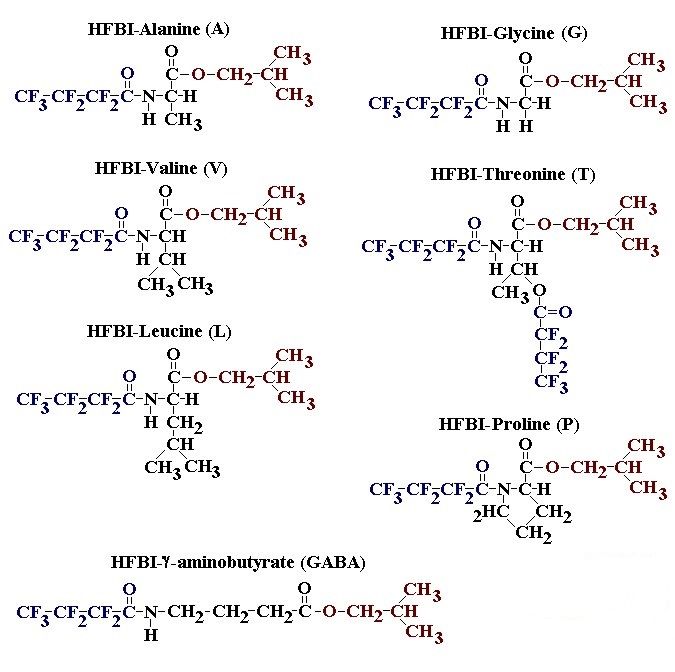 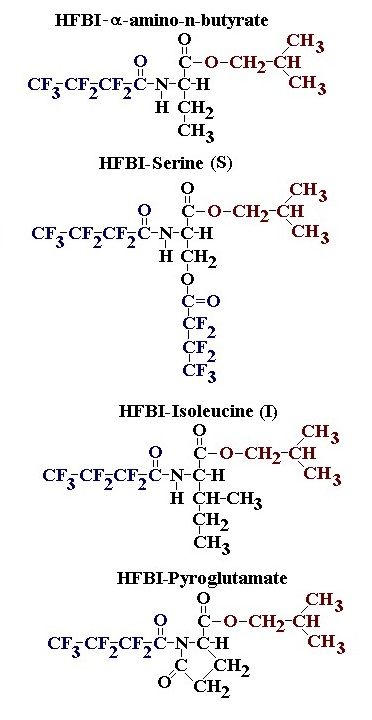 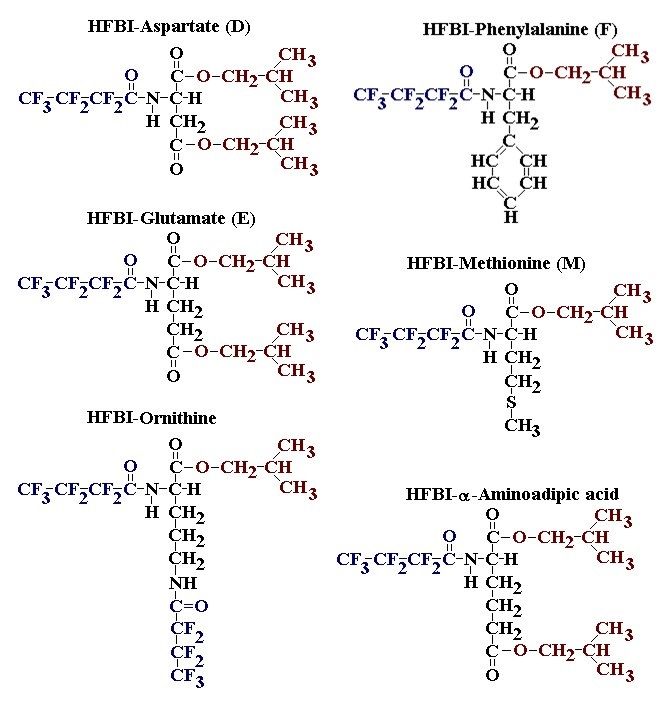  *Electron Ionization Mass Spectra*. The following images summarize the electron ionization (ei) mass spectra of a selected number of amino acids (including the amino acid standard mixture (Std), when present) through the 9 h time-course of labeling with <sup>13</sup>CO<sub>2</sub>: 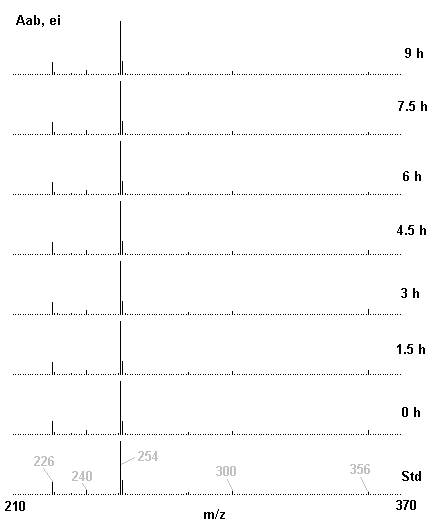 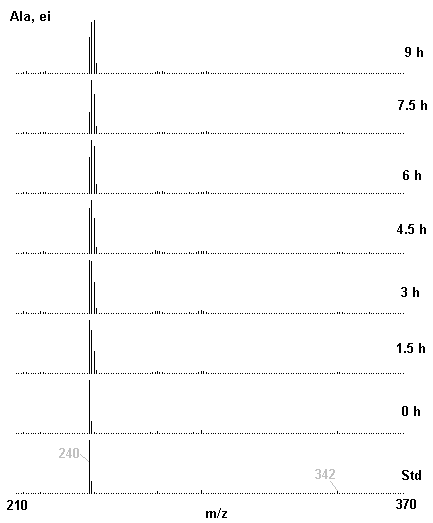 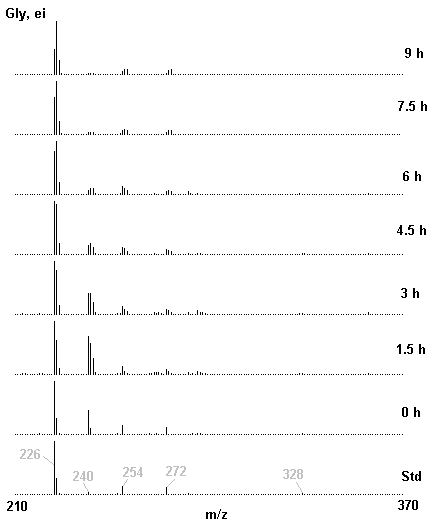 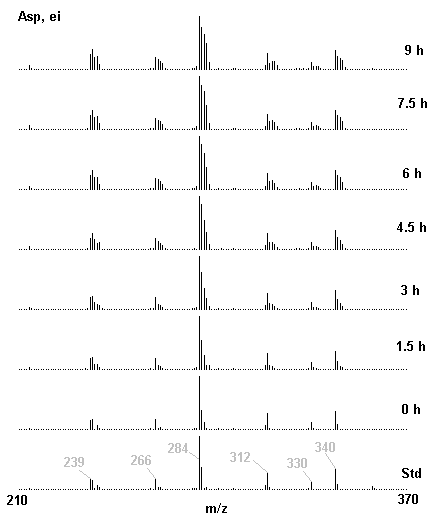 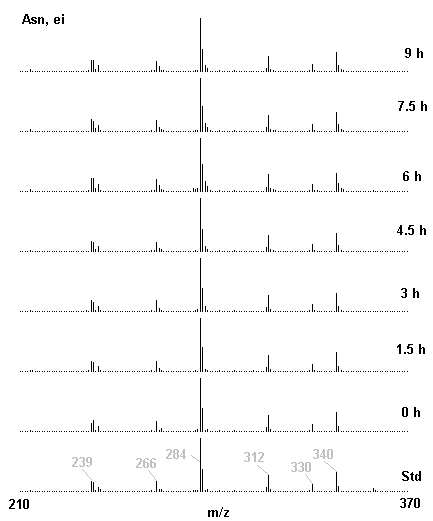 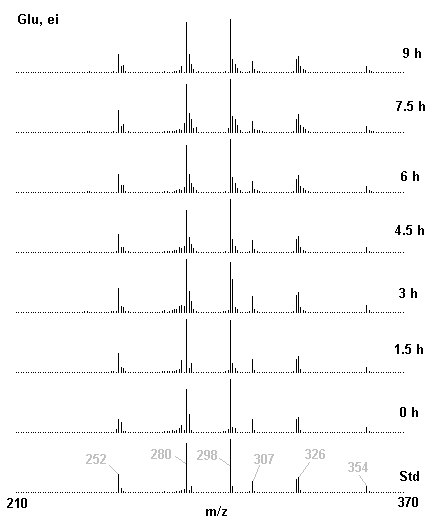 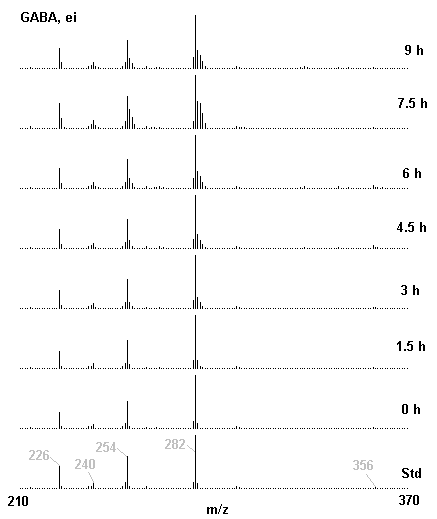 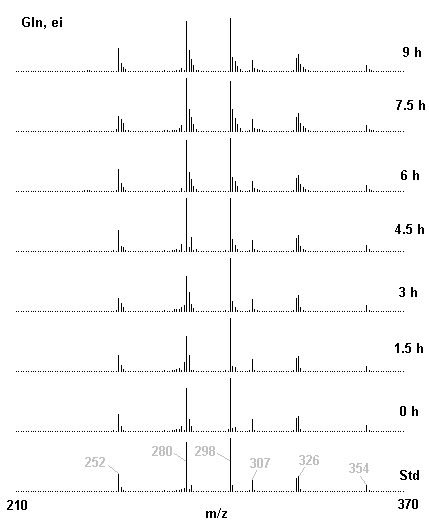 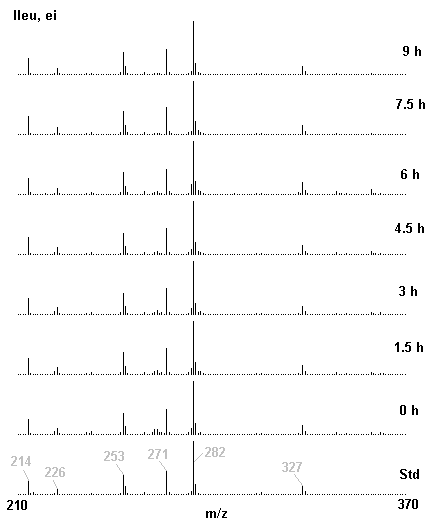 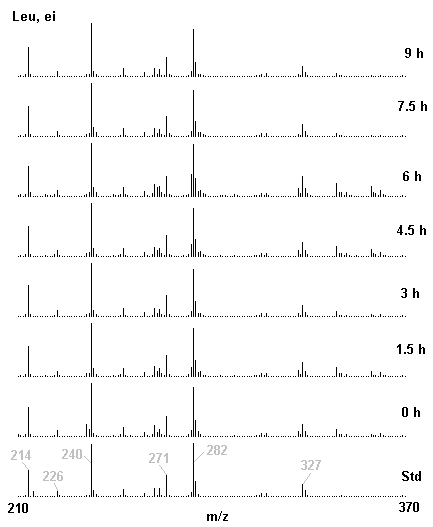 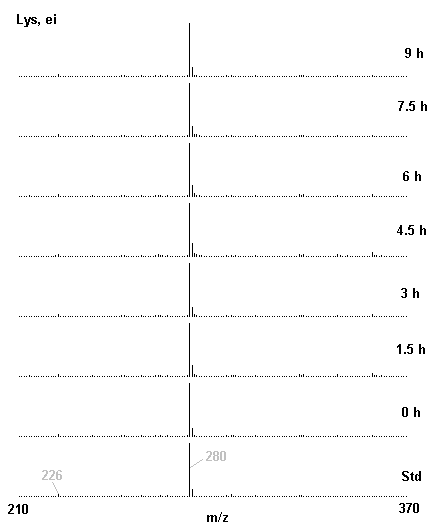 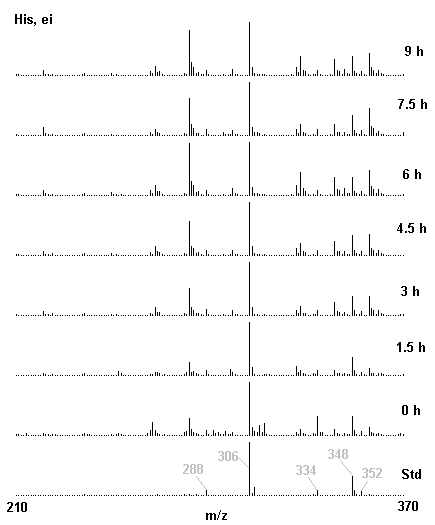 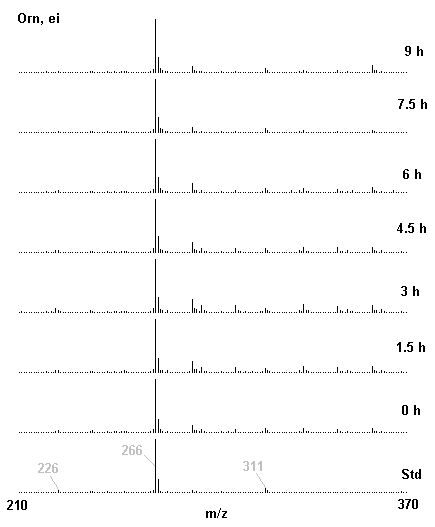 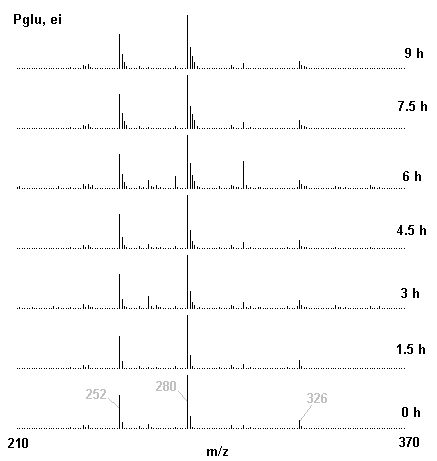 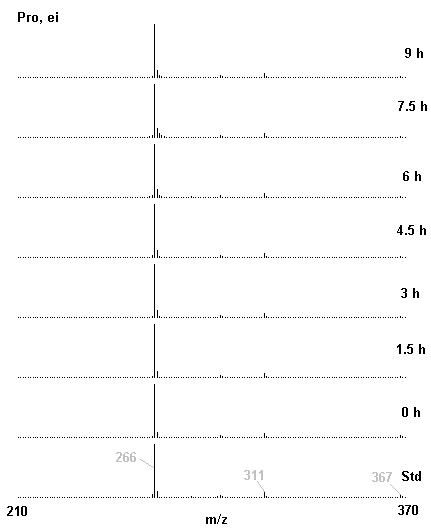 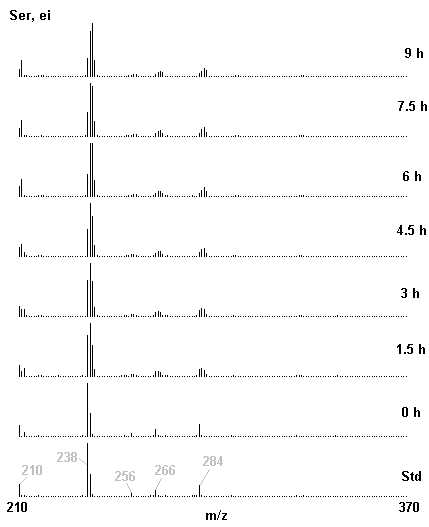 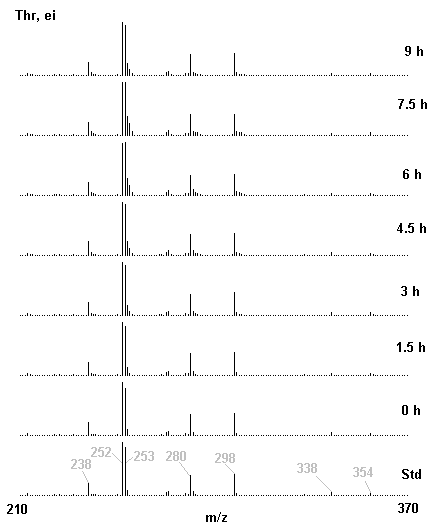 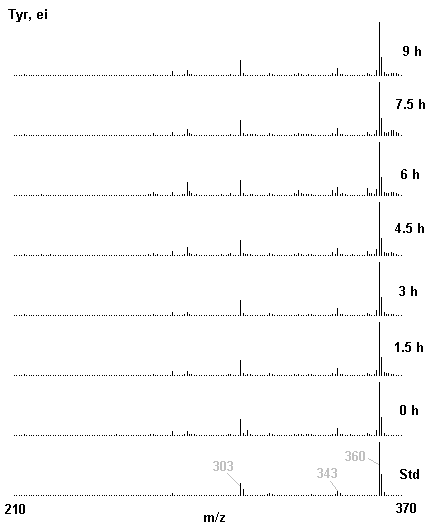 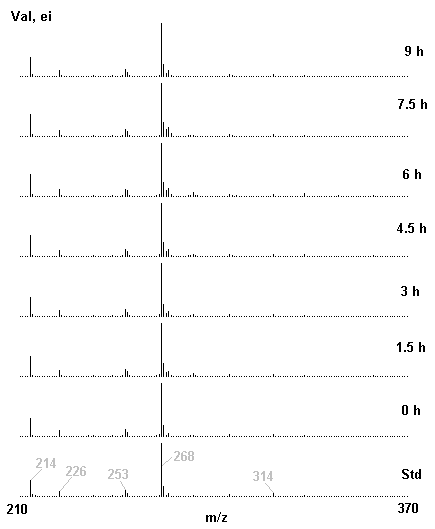 Clearly some amino acids (e.g. Ala, Gly, Ser, and Asp) acquire substantial label (i.e. their masses shift upward) early during the labeling period, suggesting that flux through these amino acids is relatively high. Others (e.g. Val, Leu, Ileu, Pro and His) acquire little label, suggesting that flux through these amino acids is relatively low. As expected, the internal standard Aab showed no isotopic labeling. Similarly the internal standard Aad remained unlabeled (not shown). The quantitative interpretation of these pool size and labeling data will be the subject of my next article. **References** [Brunk, D.G., Rhodes, D. Amino acid metabolism of *Lemna minor* L. III. Responses to aminooxyacetate. Plant Physiol. 87: 447-453 (1988)](http://www.plantphysiol.org/content/87/2/447.long) [MacKenzie, S.L., Tenaschuk, D. Gas-liquid chromatography of N-heptafluorobutyryl isobutyl esters of amino acids. J. Chromatog. 97: 19-24 (1974)](https://www.ncbi.nlm.nih.gov/pubmed/4421841) [Mayer, R.R., Cherry, J.H., Rhodes, D. Effects of heat shock on amino acid metabolism of cowpea cells. Plant Physiol. 94: 796-810 (1990)](http://www.plantphysiol.org/content/94/2/796.long) [Morgan, J.A., Rhodes, D. Mathematical modeling of plant metabolic pathways. Metab. Eng. 4: 80-89 (2002)](https://www.sciencedirect.com/science/article/pii/S1096717601902113) [Rhodes, D., Deal, L., Haworth, P., Jamieson, G.C., Reuter, C.C., Ericson, M.C. Amino acid metabolism of *Lemna minor* L. I. Responses to methionine sulfoximine. Plant Physiol. 82: 1057-1062 (1986a)](http://www.plantphysiol.org/content/82/4/1057.long) [Rhodes, D., Handa, S., Bressan, R.A. Metabolic changes associated with adaptation of plant cells to water stress. Plant Physiol. 82: 890-903 (1986b)](http://www.plantphysiol.org/content/82/4/890.long) [Rhodes, D., Hogan, A.L., Deal, L., Jamieson, G.C., Haworth, P. Amino acid metabolism of *Lemna minor* L. II. Responses to chlorsulfuron. Plant Physiol. 84: 775-780 (1987)](http://www.plantphysiol.org/content/84/3/775.long) [Rhodes, D., Myers, A.C., Jamieson, G.C. Gas chromatography-mass spectrometry of N-heptafluorobutyryl isobutyl esters of amino acids in the analysis of the kinetics of [<sup>15</sup>N]H<sub>4</sub><sup>+</sup> assimilation in *Lemna minor* L. Plant Physiol. 68: 1197-1205 (1981)](http://www.plantphysiol.org/content/68/5/1197.long) [Rhodes, D., Rich, P.J., Brunk, D.G. Amino acid metabolism of *Lemna minor* L. IV. <sup>15</sup>N-Labeling of the amide and amino groups of glutamine and asparagine. Plant Physiol. 89: 1161-1171 (1989)](http://www.plantphysiol.org/content/89/4/1161.long) [Siezen, R.J., Mague, T.H. Gas-liquid chromatography of the N-heptafluorobutyryl isobutyl esters of fifty biologically interesting amino acids. J. Chromatog. 130: 151-160 (1977)](https://www.ncbi.nlm.nih.gov/pubmed/853073)
👍 smelly, qiyi, marketstack, hr1, liberviarum, black-man, astronomyizfun, remlaps2, cub2, remlaps, sensation, davidrhodes124, aboutcoolscience, micro24, procrastilearner, tensor, vinamra, humor-room, ladeyche, bercyt, pilyug, dcdc, ribbitingscience, mobbs, steemstem, lemouth, anarchyhasnogods, flurgx, joseg, felixrodriguez, simplifylife, mayowadavid, erodedthoughts, enzor, robotics101, tristan-muller, sco, adetola, jlmol7, mittymartz, hadji, rbm, rionpistorius, kingabesh, ajpacheco1610, anyes2013, simplicitytech, effofex, count-antonio, de-stem, ari16, temitayo-pelumi, beautyinscience, justtryme90, lafona-miner, monie, deutsch-boost, doctor-cog-diss, mountain.phil28, croctopus, jaycem, biomimi, funster, borislavzlatanov, fredrikaa, abigail-dantes, thevenusproject, dysfunctional, the-devil, foundation, dna-replication, himal, curie, star-vc, locikll, lamouthe, markangeltrueman, tantawi, aboutyourbiz, howtostartablog, zacherybinx, gambit.coin, leczy, phogyan, birgitt, spectrums, motivatorjoshua, xanderslee, musicayfarandula, thedrewshow, speaklife, victoryudofia, anwenbaumeister, jacalf, karyah1001, rachelsmantra, crescendoofpeace, geekis, randomwanderings, kimp0gi, chimtivers96, hendrikdegrote, makrotheblack, nitesh9, skycae, esaia.mystic, kekegist, strings, loydjayme25, dyancuex, infinitelearning, wandersells, karolisp, dashfit, pacokam8, kerriknox, vadimlasca, smafey, laritheghost, emirfirlar, raoul.poenar, fibrefox, ertwro, muliadi, mathowl, debbietiyan, ylich, chillingotter, ikeror, sethroot, lrsm13, mahdiyari, saunter, runningman, bobdos, paulthebeloved, rasamuel, aaronteng, phaazer1, qberry, juanjdiaz89, onethousandwords, cerventus, saunter-pl, kushed, steem-hikers, jpmkikoy, jgpro, jamhuery, meetmysuperego, caitycat, etaletai, soundworks, churchboy, victorcovrig, qanon1111, boynashruddin, misterakpan, clweeks, digitalpnut, steempeninsula, loudetteiam, justinmullet, ejhaasteem, opheliapoe, mountainjewel, faithcalls, tasjun, engmi, carolynseymour, ghostgtr, kendallron, steem.curator, ewuoso, fischkopp, zeeshan003, pangoli, gra, rockeynayak, physics.benjamin, rjbauer85, serylt, sakura1012, lenin-mccarthy, positiveninja, nedspeaks, kryzsec, wisewoof, chloroform, sci-guy, ugonma, amavi, dexterdev, dber, akeelsingh, tormiwah, suravsingh, gentleshaid, blessing97, kenadis, imamalkimas, carloserp-2000, wildmanhowling, the-naked-geek, jdc, wdoutjah, mindscapephotos, damzxyno, spederson, thinknzombie, helgapn, massivevibration, onartbali, happychild, benleemusic, ntowl, torico, coloringiship, jaeydallah, bennettitalia, aamin, steepup, apteacher, vanessahampton, aehiguese, delph-in-holland, theunlimited, thatterrioguy, cordeta, fidelpoet, alinabarbu, csusbgeochem1,