The Tropical rainforest (Part 3)
environment·@steemjeet·
0.000 HBDThe Tropical rainforest (Part 3)
<h2>The importance of phylogeny</h2> There can be no doubt that a species is strongly influenced in many of its characters by its antecedents. We must therefore question whether the patterns ecologists see when they compare species, such as trees in the tropical rain forest, are not mostly reflections of phylogenetic relationships rather than recent ecological adaptations. Can we estimate the degree of ‘phylogenetic constraint’ on the ecology of species? Or can we control for the influence of phylogeny when we design ecological experiments or observations? The answer to the first question is a qualified ‘yes’. Techniques exist that attempt to partition interspecific variation into ‘ecological’ and ‘phylogenetic’ components, or at least filter out the phylogenetic effects as error in the statistical model. Two main approaches are available (Gittleman & Luh 1992). Autoregression techniques can be used that partition variance to different phylogenetic distances within the data set,generally by using either nested analysis of variance (ANOVA) or spatial statistics (Moran’s I). The alternative is phylogenetically independent contrasts (PIC), which overcome the statistical dependence of species by restricting comparisons to adjacent branch pairs on the phylogenetic tree. The major problem with applying these techniques is that they require a phylogeny from which to work, although statistical ways round this have been suggested (Martins 1996). Only recently have detailed phylogenetic analyses of the higher plants become available, and these generally stop at the family level,although more are becoming available within orders and families. These phylogenies are mostly based on DNA sequences and rarely include more than one gene, often with no more than the barest minimum of sampling per taxon. Phylogenies drawn from data of this sort suffer from many interpretive problems (outlined by Donoghue & Ackerly 1996) and at present cannot be taken as definitive. It is not surprising therefore that some ecologists have argued that phylogenetic knowledge is still too fragmentary to consider any meaningful attempt at partitioning ‘ecological’ and ‘phylogenetic’ factors in comparative analyses of tropical trees; see, for example, Hammond & Brown (1995). Most others attempting phylogenetic analyses of comparative data for tropical trees have resorted to the use of classical taxonomies for the base phylogeny (Kelly & Purvis 1993; Kelly 1995; Metcalfe & Grubb 1995; Grubb & Metcalfe 1996; Osunkoya 1996). Most of these studies claim to use PICs in their comparative analysis, but this is not strictly true. PIC is correctly a comparison of sister, ultimate branches on the phylogeny. Instead these tropical studies contrast species within the same taxon, usually species in the same genus, but extending as far as species in the same order in one case (Metcalfe & Grubb 1995). This approach is equivalent to assuming a phylogeny for the taxon concerned of instantaneous radiation from the root species to all those currently extant, hardly a plausible evolutionary pathway.Another approach has been to assume that higher taxonomic ranks should possess a greater statistical independence from phylogeny than the species level, so means within genera (see, for example, Grubb & Metcalfe 1996) or families (Baker et al. 1998) are used as the basis for comparison. What does the ‘ecological’ component of a character trait represent? If assessed by means of an accurate phylogeny, then it will represent the proportion of the character trait acquired subsequent to the last speciation event, which we can assume has a major adaptive element, particularly if repeatedly observed over many PICs. This does not mean that the ‘phylogenetic’ component is not adaptive. If a particular character state evolved in an ancestral species as an adaptation to its environment then it may still fulfil its adaptive role in the daughter species. However, phylogeny may constrain the range of variation that a descendant can exhibit, and what was once adaptive may no longer be, but merely represent the inertia of evolutionary change. We have returned to the thorny problem of the question of proof in the study of adaptation.Does phylogenetic analysis get us any further in our understanding? To date the only area of research in tropical tree ecology where phylogenetic analysis has been applied more than once is the question of the relation between seed size and shade tolerance (Kelly & Purvis 1993; Kelly 1995; Metcalfe & Grubb 1995; Grubb & Metcalfe 1996). Studies of a range of tropical tree species at any particular site generally show that those believed to be light-demanding for regeneration have significantly smaller seeds on average than those that are shade-tolerant (Foster & Janson 1985; Metcalfe & Grubb 1995; Hammond & Brown 1995). But phylogenetically controlled analyses have failed to find significance, or even found larger seed size in light-demanders (see, for example, Fig. 1.4). This implies that there is a strong phylogenetic component to seed size that includes both inertia and inherited adaptation. The absence of the predicted trend in the ecological component may indicate that seed size is less adaptive than previously thought. One of the problems with comparative analyses conducted thus far on tropical trees is that the data were usually collected with the aim of being as thorough as possible: as many species as were available were included. However, when phylogenetic effects are partitioned many of the data have little weight in the analysis because the species are too isolated phylogenetically from other species of contrasting ecology to provide much information.It would be more efficient to predetermine which species were to be contrasted. There has been an increasing frequency of published studies of congeneric species, which may be one way of efficiently tackling the conundrum of phylogenetic influence on ecology. The detailed investigations of the regeneration of Shorea section Doona in Sri Lanka. 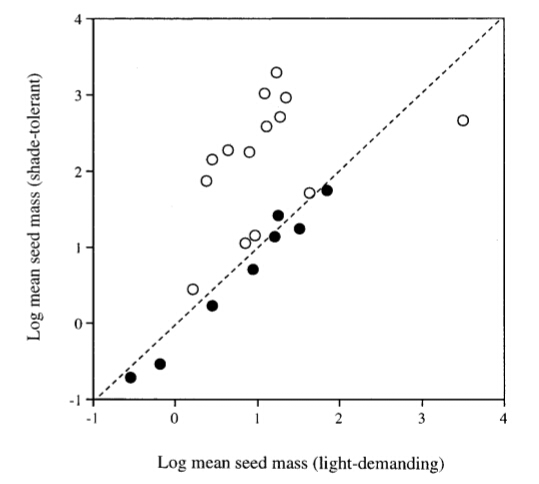 <h6>Figure 1.4 Scattergram of the mean seed dry mass values (mg) of shade- tolerant taxa relative to those of light-demanding taxa in phylogenetically controlled comparisons. Open and closed circles indicate genera within families and species within genera, respectively. After Grubb & Metcalfe (1996).</h6> (Ashton 1995; Ashton etal. 1995; Gunatilleke et al. 1996a,b, 1997) and those of eleven sympatric and closely related species of Macaranga in Sarawak (Davies et al. 1998; Davies 1998; Davies & Ashton 1999) stand out as good examples. <h3>The ecological classification of tropical rain-forest trees</h3> An ecological classificatory system for tropical trees is needed in order to organise our information about the species concerned. The systematisation of knowledge is one of the most important activities of science.A logical organisation efficiently condenses information into a series of generalisations, and allows predictions to be made about members of groups recognised by the system. Without such a system, ecology is reduced to natural history employing the scientific method. This is not meant to denigrate natural history. Natural history provides many of the observations that inspire the formulation of hypotheses, and data by which these hypotheses are tested, but it is science that organises the corpus of natural-history knowledge. Diversity is the main hurdle faced in designing an ecological classification of tropical trees. The sheer number of tropical tree species means we have only just begun to have any information at all about a very large number of species. Ghana, in West Africa, is probably the only substantial tropical area where all the tree species have been considered ecologically (Hawthorne 1995). Therefore there is an inherent danger that we try to force new data into our pre-existing framework, when maybe we should re-examine the system of organisation we are using. There are a variety of approaches that can be used to erect an ecological classification system. In classifying the classifications, as it were, one dichotomy might be between systems emphasising groups, and those that emphasise the means of differentiating the groups. Any system of classification has to include both elements, but it may be that the groups are defined by arbitrary points along the axes of differentiation, making the groups less important than the axes. This is the commonest state of affairs in plant ecological classification systems, because discrete groupings of species rarely occur. Axes of differentiation are often defined in terms of the characteristics of species that are thought to lie near the ends of each axis; for instance shade tolerance is usually defined by comparing its end points. Another distinction might be between those systems derived from a priori data analysis and those proposed from general observations without recourse to quantitative data. Most of the classificatory systems proposed for tropical trees are of the second type. A good example is Swaine & Whitmore’s (1988)pioneer versus non-pioneer classification of tropical trees. This was based on a wide range of observations made by many different foresters and ecologists,but did not involve any statistical tests of either the character correlations described or the group differentiation proposed. The alternative method of gathering data and performing multi-variate analysis to investigate trends and discontinuities has less commonly been done, although examples are available (see, for example, Condit et al. 1996a; Gitay et al. 1999). More studies along these lines will be of value to improving our current classification systems.
👍 rk786, bubbleburst, originalworks, ohreally, gotsk, shablinscky, yry64, evgenijpax, frenertui, samkunova, break9, ijenirina, freyman, michelnilles, steemjeet, ornima, banjo, minnowsupport, pharesim, barrie, endaksi1, edrivegom, gomeravibz, jhermanbeans, steemprentice, valth, pomperipossa, arjane, decibel, ilvacca, jhagi.bhai, gindor, whatamidoing, beng05, timbalabuch, qwasert, pusteblume, cryptohustler, myday, gamerveda, nesbitt, bluchr, a2jimenez, stephen.king989, lastminuteman, numpypython, taica, tinashe, tradewonk, drotto, masudrana, as-i-see-it, pradipbhattarai,