How to Earn a Nobel Prize Part 2: George P. Smith and Phage Display
steemstem·@tking77798·
0.000 HBDHow to Earn a Nobel Prize Part 2: George P. Smith and Phage Display
To celebrate SteemSTEM’s launch of their new website (https://steemstem.io), I’m writing a series of posts on the recently announced 2018 Nobel Prize in Chemistry. This year’s prize was awarded to Frances H. Arnold for pioneering the first directed evolution of enzymes, and to George P. Smith and Sir Gregory P. Winter for developing phage display [1](https://old.nobelprize.org/che-press.pdf?_ga=2.87232604.535716584.1538560295-715396971.1538560295). The award was a major story in a bunch of news outlets, but I found most of the pieces were scant on details. They only gave a cursory overview of the techniques that the scientists spent a major portion of their lives developing. Fortunately, @SteemSTEM has given me the platform to remedy that problem. Each entry in this series will focus on one of the scientists’ contributions, try to explain that contribution and show why the scientist deserves this prize. Today’s focus: George P. Smith and Phage Display. # Who is He? <div class="pull-right"> <center> https://cdn.steemitimages.com/DQmPxc2N188LpEkL2X8m8J4hiutipCkqDydETiVAR6aLGmD/image.png <sup> <br> George P. Smith (2). <br> He genuinely looks like a happy guy :) </sup> </center> </div> George P. Smith has an impressive, albeit unexciting academic career on paper. He got his Ph. D in Bacteriology and Immunology at Harvard University, took a postdoc at the University of Wisconsin, and then became a professor at the University of Missouri in 1975 [3](https://en.wikipedia.org/wiki/George_Smith_(chemist)). For the next 43 years, he continued to be a faculty member at the University of Missouri and is now a Professor Emeritus. This position usually refers to a semi-retired professor that is still given university resources to pursue his or her research interests, but doesn’t have the responsibilities of a full professor (all professors, whether they know it or not, strive to become a Professor Emeritus and then die at the bench). George Smith’s current research focuses on using his phage display technology to visualize cancer, and he still publishes every few years. All in all, George P. Smith’s career might have been unremarkable if not for the magic of sabbaticals. A sabbatical is a paid leave of absence for professors, usually lasting a year and taken once for every seven or so years worked. Back in the pre-internet era, this was a key opportunity for academics to broaden their horizons. They could learn first-hand from other specialized labs and bring that specialized knowledge to their university. Sabbaticals are a bit less important now that we can share information so easily, but they still provide an excellent opportunity to work with resources and experts that might not be available at a professor’s home university. Some professors blow the opportunity off, but George Smith knew exactly what he wanted to do. He secured a spot in Robert Webster’s lab at Duke University which had the equipment and know-how accomplish his goals and got to work [4](https://medschool.duke.edu/about-us/news-and-communications/med-school-blog/nobel-prize-winner-george-p-smith-completed-early-award-winning-research-duke). You could argue that George P. Smith had the greatest sabbatical in history as he used it to develop a revolutionary technology that cemented his position in science history. Oddly enough, this isn’t the first George Smith to win a Nobel prize. In 2009, George E. Smith won the 2009 Nobel Prize in Physics for helping develop digital imaging technology integral to modern digital cameras (https://www.nobelprize.org/prizes/physics/2009/smith/facts/). This isn’t important, but it made researching this guy’s bio a heck of a lot harder so I needed to vent about it. # What did he do? The technology in question is known as phage display, a technique designed to improve the utility of directed evolution. The process of directed evolution was developed by Frances Arnold, the subject of the previous post in this series. In brief, directed evolution was a rapid way to generate thousands of protein variants, and completely changed the way scientists could improve upon proteins. However, it still had a major weakness in that it didn’t provide an easy way to analyze the created protein variants. In Dr. Arnold’s original paper, she used a specialized protein with a unique method of assessing activity, but that method doesn’t apply to proteins in general. In fact, most proteins stay inside a cell and are difficult to analyze without breaking open the cells they’re in. Not only is this time consuming, but it’s hard to keep track of the cell that a particular protein variant came from. Dr. Smith circumvented this problem by genetically engineering bacterial viruses (known as phages) to express the proteins of interest on their outer coats allowing for them to be easily assessed or enriched. It was called phage display because the phage is exposing or “displaying” these proteins to the external environment. His initial work on developing this technology can be found in the science publication *Filamentous Fusion Phage: Novel Expression Vectors That Display Cloned Antigens on the Virion Surface* [5](http://science.sciencemag.org/content/228/4705/1315) and this article does a great job of laying out the groundwork for what would become known as phage display. Phage display works by fusing a gene of interest into the middle of a viral gene that normally exports to the surface of the phage. Dr. Smith choose filamentous phage gene III which has a distinct signal peptide sequence. <center> 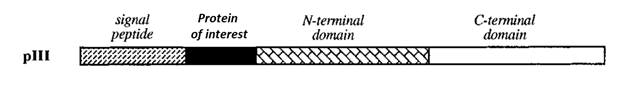 <sup> Diagram of the protein of interest fused within the filamentous phage protein III [6](http://www.biosci.missouri.edu/SmithGP/PhageDisplayWebsite/PetrenkoSmithChemReviews.PDF). </sup> </center> When this signal peptide is synthesized, the virus exports the attached protein to the outer coat of the phage. When the protein of interest is fused between the signal peptide and the rest of the protein, it will wind up on the outside of the virus with the rest of the protein tethering it to the outer coat. <div class="pull-left"> <center> https://cdn.steemitimages.com/DQmY3Z1r3V16QepiFrTPFhHwrMK4Ly57c18wDoj7TABHyMQ/image.png <br> <sup> The protein of interest (hatched circles) is exported to the outside of the phage and stays attached by a portion of the original protein (white circles) (6). </sup> </center> </div> The protein will stay firmly attached to the phage, but can be exposed to external stimuli. This exposed-protein attached to a virus arrangement opens up a lot of opportunities for scientists. The most obvious to Dr. Smith (and generally the most effective use of this technology) was affinity purification. That is, scientists can use phage display to isolate proteins that bind to a specific substrate of interest (like an antibody), then propogate the phage to amplify the proteins that bind the strongest. The article makes it clear from the start that this is a proof-of-concept paper. Dr. Smith’s main goal is to show that his idea of phage display works in practice, so he started with a model protein. That protein is the EcoRI endonuclease [7](https://www.neb.com/products/r0101-ecori#Product%20Information). This is a well-studied protein that cleaves a specific DNA sequence (GAATTC to be exact). More importantly, antibodies specific to this protein are readily available so affinity purification is relatively easy. Dr. Smith artificially created a mixture in which phages displaying EcoRI were vastly outnumbered by a control phage (phage m13mp3). The specifics of why the control phage was chosen is a bit beyond the scope of this article, but suffice to say it can easily be quantified and can be distinguished from the EcoRI displaying phage. The exact steps of the experiment were as follows: Dr. smith adds a few EcoRI phages in a big mixture of control phage and spreads the mixture onto a petri dish with affixed antibodies specific to EcoRI. The mixture is allowed to incubate so that all the EcoRI displaying phages bind to the affixed antibodies. The plate is then washed multiple times which removes the control phage while the bound EcoRI displaying phages remain. Finally, the phages that remain bound after multiple washes are eluted and the amount of control phage and EcoRI phage relative to the initial phage input are recorded. <center>  <sup> Affinity purification of phage with an EcoRI gene fusion from control M13mp8 phage [5](http://science.sciencemag.org/content/228/4705/1315). </sup> </center> In the results shown above, you can see that the washes removed almost all the added phage, with less than 0.5 % of the total phage added remaining. However, the amount of EcoRI phage that remained was vastly higher than the amount of control phage remaining. In this particular experiment it was enriched by 3400. Thus, the affinity purification was successful which confirms that Dr. Smith’s concept of phage display works in practice. # Why does this deserve the Nobel Prize? <div class="pull-right"> <center> https://cdn.steemitimages.com/DQmZbCkfvHJWahVQYpfME3JSNox37xfrAzWDutWgkRHsuRS/image.png <sup> <br> Diagram showing how phage display can be used to isolate and analyze the red protein of interest (8). </sup> </center> </div> This novel technique is important because it vastly expands the applications for directed evolution. As I said before, Dr. Frances Arnold’s directed evolution technique was limited because it didn’t provide a way to assess protein activity. Phage display provided a viable solution to this limitation. Directed evolution could be used to create thousands of gene variants, which could then be inserted into thousands of phages. Those phages would display the protein variants on their coats, and a substrate could be used to capture the variants that were most useful. Dr. Smith specifically calls out antibodies as a useful tool for capturing, but many other options are available. For example, if you wanted to engineer a protein that binds to a specific DNA sequence, you can affix that DNA sequence to a surface and use it to capture phages displaying only the protein that binds to the DNA. You can then let the phage infect bacteria, reproduce, and ultimately isolate that DNA to determine the exact sequence of the protein and have a DNA insert with which to perform further transformations. It’s also important to note here that Dr. Smith didn’t stop his work on phage display with this single paper. He continued to push this technology forward and make it more and more useful. In this sense, the Nobel prize recognizes a lifetime of achievement rather than just one lucky finding during a sabbatical. There are many, ***many*** applications for phage display. From basic research purposes like receptor identification to more applied functions like drug discovery or vaccine development [6](http://www.biosci.missouri.edu/SmithGP/PhageDisplayWebsite/PetrenkoSmithChemReviews.PDF). Next week, I’ll be delving into one major way in which the technology of phage display was adapted to create antibodies for therapeutic uses. This work was the product of Gregory P. Winter, the third and final recipient of this Nobel prize. # References (1) https://old.nobelprize.org/che-press.pdf (2) https://biology.missouri.edu/people/?person=94 (3) https://en.wikipedia.org/wiki/George_Smith_(chemist) (4) https://medschool.duke.edu/about-us/news-and-communications/med-school-blog/nobel-prize-winner-george-p-smith-completed-early-award-winning-research-duke (5) http://science.sciencemag.org/content/228/4705/1315 (6) http://www.biosci.missouri.edu/SmithGP/PhageDisplayWebsite/PetrenkoSmithChemReviews.PDF (7) https://www.neb.com/products/r0101-ecori#Product%20Information (8) https://en.wikipedia.org/wiki/Phage_display # Images All images were taken either directly from the publication referenced or labeled for reuse on Google Images. George P. Smith’s photo was taken from the University of Missouri’s faculty page. If any image owner has an issue with this article, please contact me and I will address the issue. https://steemitimages.com/0x0/https://i.imgur.com/WhYkkh9.gif
👍 tfcoates, mariobarrientos, nfc, merlion, emrebeyler, angelinafx, bitok.xyz, whitebot, steemchoose, bloom, vjap55, sbi7, liberviarum, scienceangel, sensation, alexs1320, steemstem, lemouth, dna-replication, alexzicky, robotics101, effofex, kevinwong, alexander.alexis, fancybrothers, jpederson96, tanyaschutte, felixrodriguez, espoem, enzor, eleonardo, tristan-muller, fejiro, sco, adetola, rharphelle, jlmol7, real2josh, traviseric, rionpistorius, heajin, kingabesh, ajpacheco1610, charitybot, lianaakobian, anyes2013, dedesuryani, count-antonio, de-stem, ari16, temitayo-pelumi, alexworld, frost1903, synick, beautyinscience, osariemen, kingnosa, curie, kryzsec, helo, suesa, ludmila.kyriakou, howo, tsoldovieri, esteemguy, mr-aaron, fbslo, erikkun28, delin.english, terrylovejoy, olajidekehinde, flugschwein, francostem, emmanuel293, derbesserwisser, greatwarrior79, outtheshellvlog, michaelwrites, deholt, ibk-gabriel, osakuni, reyvaj, purelyscience, mitaenda, drmoises, scottallen, herbayomi, swapsteem, javier.dejuan, leysonrye0724, samminator, abigail-dantes, nicola71, kenadis, gentleshaid, mathowl, dexterdev, alexdory, deusjudo, lesmouths-travel, kingabesh1, stem.witness, tombstone, mrwang, arconite, mountain.phil28, nurhayati, jesusj1, monie, mondodidave73, tajstar, jaycem, charitymemes, doctor-cog-diss, steemzeiger, biomimi, funster, wstanley226, suasteguimichel, testomilian, allhailfish, andiblok, locikll, aboutyourbiz, corsica, gambit.coin, kingswisdom, gabox, irishcoffee, kerry234, iptrucs, votetanding, speaklife, darkiche, dragibusss, galam, chimtivers96, elith, vact, ihsan19, maxruebensal, wandersells, skycae, strings, loydjayme25, sissyjill, wrpx, morbyjohn, bil.prag, anwenbaumeister, pacokam8, lrsm13, muliadi, runningman, marialefleitas, cerventus, laritheghost, aaronteng, debbietiyan, atjehsteemit, chillingotter, nitego, onethousandwords, leyla5, anna-mi, clweeks, derekvonzarovich, peaceandwar, sireh, operahoser, jpmkikoy, beladro, niouton, hendrikdegrote, carolynseymour, saintopic, gotgame, emdesan, digitalpnut, kendallron, tasjun, qberryfarms, joanpablo, raymondspeaks, moksamol, thatsweeneyguy, jayna, g0nr0gue, indy8phish, paddygsound, joendegz, mhm-philippines, anikekirsten, stahlberg, bestsmiles, jembee, shookriya, onethousandpics, cryptoisfun, hansmast, getrichordie, creatrixity, didic, bavi, hardaeborla, mahmudulhassan, ashfaaaq, juanhobos, bitson, joshglen, charlie777pt, resteemer, foways, christianolu, nolasco, sweetdreams, neumannsalva, janine-ariane, drmake, tomatom, dolphinscute, mrday, technotroll, avizor, romanleopold, cosmophobia, call-me-howie, synthtology, diana.catherine, akumar, catalincernat, m1alsan, adamzi, niko3d, ameliabartlett, elfranz, joelagbo, kookyan, galione, disruptivas, deividluchi, clement.poiret, kiikoh, mrunderstood, praditya, predict-crypto, gangstayid, dbzfan4awhile, gabrielatravels, hetty-rowan, vilda, giddyupngo, idkpdx, alexa.creates, momimalhi, dikkie, raghao, rhethypo, chickenmeat, tuck-fheman, lola-carola, gmedley, the-eliot, mininthecity, crypto-econom1st, justasperm, juanmolina, ejgarcia, stonecoin, zerokun, semtroneum, daddywilliam, positiveninja2, lordkipas, pablocordero, slickhustler007, gauttam, venalbe, all-right, zlatkamrs, lilianajimenez, jjohnson78, warpedpoetic, adalhelm, wanasoloben, annaabi, alom8, nothingismagick, boynashruddin, jan-mccomas, riche-gould, trenz, weirdheadaches, azulear, jlsplatts, abeyaimary, hkmoon, gordon92, jordan.white306, reizak, thabiggdogg, scintillaic, melaniesaray, bestofph, shadown99, trang, kinglypalace, eu-id, homefree, sarhugo, ppss, lostplace, kanhiyachauhan, lafona-miner, circleoffriends, wdoutjah, hectgranate, tea-man, wisewoof, serylt, perpetuum-lynx, imamalkimas, steemit-bot, neneandy, supposer, justtryme90, dna-polymerase, dna-ligase, dna-helicase, dna-primase, sliding-clamp, doneliseo, clamp-loader, dna-gyrase, rna-polymerase, ribosome, jtm.support, zest, damdap, bohemian.machine, jewlzie, damzxyno, communityisyou, smacommunity, knightbjj, anthive, paradigmprospect, medicnet, thescubageek, alvin0617, mahdiyari, torico, lamouthe, eniolw, philipkavan, wackou, uceph, coquiunlimited, faustofraser, reconstitution, angelica7, raoufwilly, thomaskatan, homespun, momogrow, jiujitsu, deadcountry, landria, maski, iansart, drsensor, cherryandberry, sublimenonsense, massivevibration, happychild, lacher-prise, gribouille, benleemusic, robertbira, eroticabian, sanderdieryck, eurogee, laurentiu.negrea, hhtb, yomismosoy, orcheva, kafupraise, jbrrd, iradyjr, langford, steepup, ninyea, wargof, hillaryaa, delph-in-holland, guga34, cordeta, mrxplicit, aotearoa, apteacher, honeysara, cooknbake, brandt, christianyocte, therosepatch, oghie, bitmycoin, elohim4, minuetoacademy, danlipert, abraham10, yashshah991, faithfullwills, forestplane, indayclara, memeitbaby, titoncp, jonmagnusson, musicvoter, avesa, infamousit, wolfnworbeikood, grizzle, atomcollector, newenx, ravenmus1c, peter-ella, sussexmusicfest, anyer-quantum, isaria, kingeazi, archaimusic, bretho, lastravage, emsteemians, elamental, dinoromanelli, lillywilton, diantbi, adam-aj, skorup87, alchemicjourney, utopian-io, tamarind-juice, dominicbaitan, salimon, crytiblock, morphewq, funtown, hakansahin, ccworld, majidse, keremkoz, zephyraijunzo, fidepet, sttest2, insight-out, monkeyking1, vezo, easyimagination, adamada, mittymartz, soy-venezuelien, tking77798, iamphysical, miftah9914,